The ongoing SARS-CoV-2 pandemic has highlighted the need to understand all aspects of coronavirus biology, including their prevalence and diversity in animal hosts and the environment. Given the pressing need for greater knowledge around this topic, researchers within the Microbiome Informatics Team at EMBL’ European Bioinformatics Institute (EMBL-EBI) are repurposing existing infrastructure to identify viral genomes of the Coronaviridae family within public meta-omics datasets.
The Microbiome Informatics Team, headed by Rob Finn, is responsible for the MGnify resource, which houses one of the most extensive analysis sets for metagenomics data in the world. Utilising this resource, the team has repurposed existing workflows to generate a pipeline that detects and characterises coronaviruses from metavirome and metatranscriptomic datasets. This pipeline identified a complete SARS-CoV-2 genome from a human lung sample collected in Wuhan, China, at the start of the pandemic — demonstrating proof of concept.
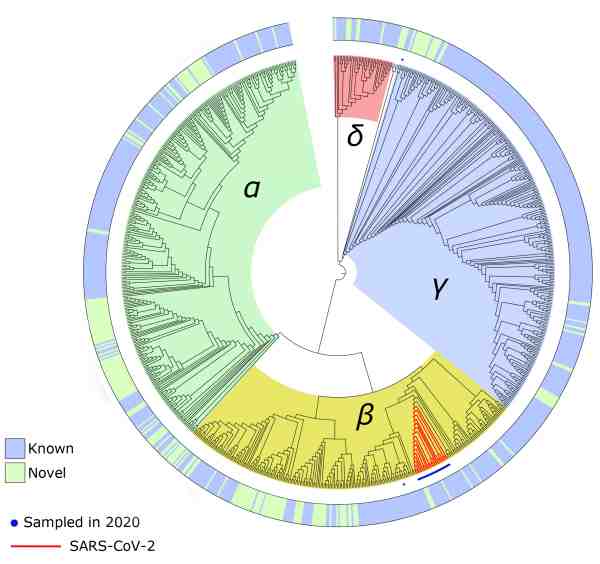
The next step for the team was to expand the search for coronaviruses in diverse environments and hosts. They plan to screen more than 70,000 metatranscriptome and metavirome datasets available in the European Nucleotide Archive (ENA), an ELIXIR Core Data Resource.
Leveraging this large dataset, alongside rich sample metadata, expands the knowledge of coronavirus presence in the environment and provides a comprehensive reference point for future genomic, epidemiological and functional studies.
However, research projects, such as those undertaken by Rob Finn and his team, are highly dependent on rich metagenomic sample data and sustained funding of research infrastructures.
The global response to SARS-CoV-2 has been swift and extensive; but it has also exposed many issues, including the perils of unsustainable infrastructures and neglected metadata. For instance, in June, the Genomic Standard Consortium (GSC) published an article in Scientific Data highlighting this issue, with the authors stressing the need for standardisation methods to reuse genomic data.
These two factors, high-quality metadata and the ability to repurpose tools, allow existing networks of experts to act rapidly to urgent needs, such as pandemics. ELIXIR advocates and supports data standards and offers a sustainable research infrastructure for the repurposing of tools and services, as demonstrated by Rob and his team.