Microbiome research has grown substantially over the past decade in terms of the range of biomes sampled, identified taxa, and the volume of data derived from the samples. In particular, experimental approaches such as metagenomics, metabarcoding, metatranscriptomics, as well as other -omics are providing key information on the function, diversity, and mechanistic drivers of microbial communities.
The ELIXIR Microbiome Community aims to overcome the nonuniform practices in microbiome research by aligning standards, protocols, and methods. The community is dedicated to sharing experiences about microbiome analysis, relying on the adoption of FAIR (Findable, Accessible, Interoperable, and Reusable) principles being aware of the Digital Sequence Information (DSI) and CARE (Collective Benefit, Authority to Control, Responsibility, and Ethics) principles. Through training actions and federated data analysis, the community strives to enhance the efficiency and sustainability of bioinformatics infrastructure and resources for microbiome research. This will enable a deeper understanding of eubiotic versus dysbiotic microbial states through the development and characterization of synthetic communities that encompass the full taxonomic spectrum, including bacteria and other microbes.
Community goals
Develop standards for FAIR microbiome analyses
- Data
- Establish best practices for the submission of raw sequences data, assemblies and MAGS that can represent all biomes.
- Develop controlled vocabularies, knowledge graphs and metadata quality checks.
- Establish methods to improve metadata before and after deposition.
- Align metadata for interoperability between databases like BioSamples, and develop a training module to ensure its consistent application.
- Foster international collaborations to ensure global harmonization of e-infrastructures for microbiome research.
- Leverage data management, processing, and provenance solutions to enhance the infrastructure and resources available for microbiome research.
- Tools
- Improve annotation via bio.tools and development of EDAM terms.
- Leverage the CAMI initiative to facilitate benchmarking of tools and workflows.
- Spearhead the development of new methods and tools for diverse data analysis with projects like Odyssey, which connects molecular and geographical biodiversity data.
- Workflows
- Design and implement workflows for microbiome research, for example with projects like FAIRyMAGs, which optimizes metagenome-assembled genome workflows.
- Promoting the use of best practices and RO-Crates.
- Federated computation
- Enable the execution of MGnify pipelines in Galaxy and/or other data management workflows, and submission of results to MGnify.
- Establish routine mechanisms for federating microbiome analysis, such as RO-Crates and resources.
- Catalog
- Create a catalog of tools, workflows resources
Build capacity in microbiome analyses
- Create a catalog of training resources for microbiome analyses.
- Address knowledge gaps in generating and adopting workflows, and deliver targeted training for different microbiome communities.
- Develop training resources to increase awareness of microbiome tools and their applicability.
- Organise and participate in training events (workshops and schools) to educate researchers about microbiome best practices.
- Actively participate in and contribute to community-led projects and initiatives, such as the Galaxy Training Academy's Microbiome Analysis track.
- Build a community of trainers to enhance the skills and knowledge of researchers in microbiome analysis.
Represent the Microbiome Community in its complexity
- Evaluate needs, key datasets, data analysis approaches, omics data types, and biome-specific specialisation.
- Identify key experts involved in viral, prokaryotic, and eukaryotic analysis.
- Establish and share a strategic technical roadmap with the Communities and Platforms, highlighting key contacts.
- Identify relevant funding calls and build microbiome research informatics capacities, connecting to key experts in other omics fields such as metaproteomics.
- Develop crosstalk between distinct “biomes” area.
- Develop a connection with the industry.
- Represent the Microbiome Community at international conferences, promoting Community/ELIXIR outputs and solutions.
-
Facilitate participation to hackathons such as the ELIXIR BioHackathon to bring together microbiome researchers for collaborative problem-solving and innovation.
Community Impact
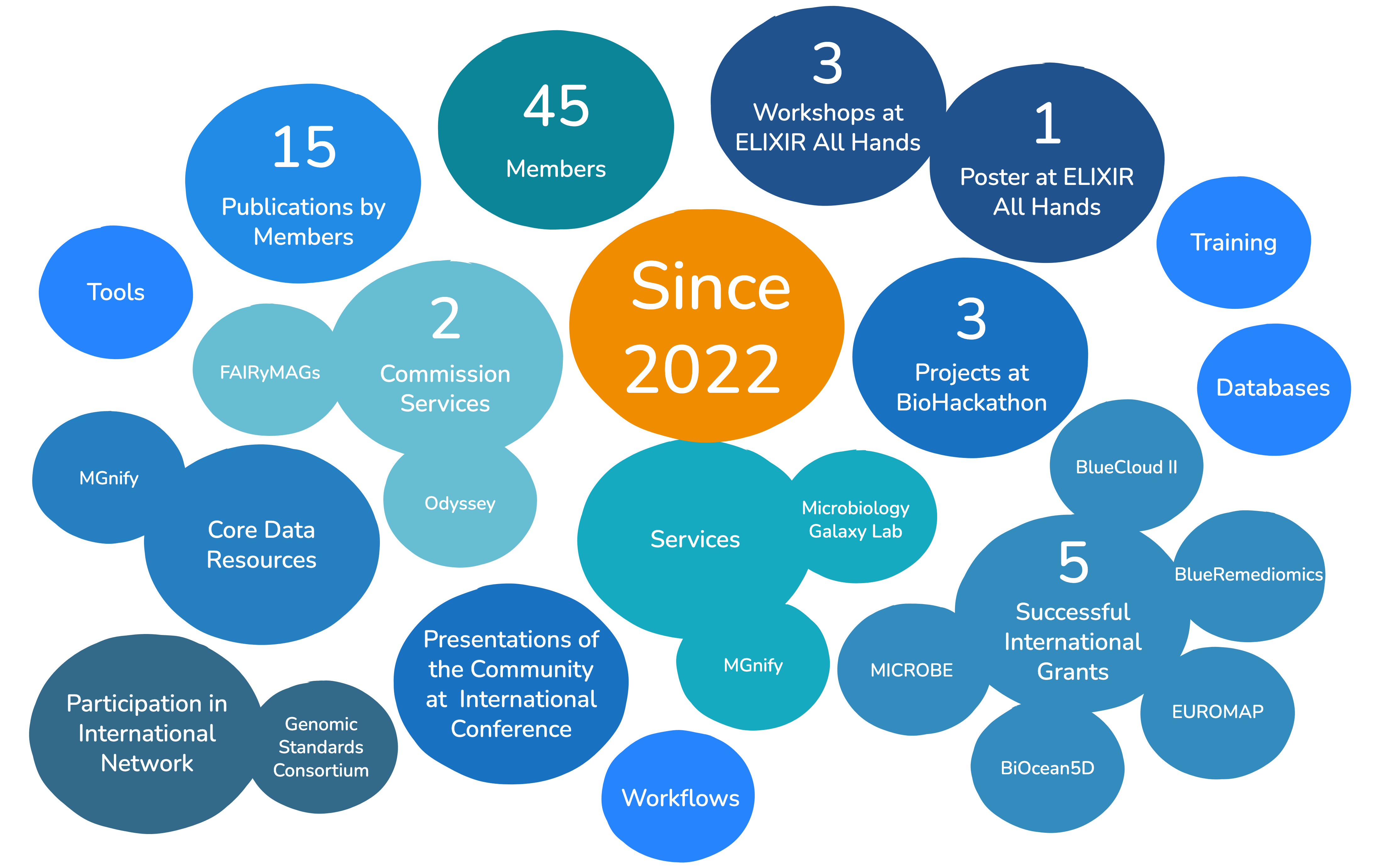
The ELIXIR Microbiome Community has made significant strides in advancing microbiome research. Key achievements include the acceptance of two ELIXIR Commissioned Services to be part of the ELIXIR 2024–28 Programme's Biodiversity, food security and pathogens Science Tier: FAIRyMAGs and Odyssey. The community's active participation in the recent ELIXIR BioHackathons and the ROcrate initiative led to the development of scalable solutions for multi-omics analyses and improved the annotation of microbiome-related tools on bio.tools.
The community has also contributed to the Microbiology Galaxy Lab manuscript, which provides a gateway to tools, workflows, and training for accessible, reproducible, and FAIR microbial data analysis. Members have participated in the Galaxy Training Academy's Microbiome Analysis track since 2021, offering tutorials and captioned recordings to the research community.
Additionally, the integration of the MGnify v5 Amplicon Workflow into Galaxy has advanced federated analysis, making it easier for researchers to perform comprehensive microbiome analyses. The community's efforts have been recognised with awards, such as the Best Poster Award at the ELIXIR All Hands meeting 2022, highlighting its impact and contributions.
The community has also been instrumental in successful grant applications, including BIOcean5D, BlueRemediomics, BlueCloud II, and MICROBE. These grants have enabled the community to leverage data management, processing, and provenance solutions, further enhancing the infrastructure and resources available for microbiome research.
Community meetings, such as the ones held in Barcelona in September 2023 and April 2025, have been pivotal in discussing objectives and connecting with other ELIXIR communities, particularly the Biodiversity community.
By focusing on these goals, objectives, and impacts, the ELIXIR Microbiome Community continues to drive advancements in microbiome research and foster collaboration among researchers across various biomes.
Leadership




- To get in touch with the Community leads, contact microbiome-coleads@elixir-europe.org
- Read the Community white paper: https://f1000research.com/articles/13-50/v2
- Microbiology Galaxy Lab
- To find out more about MGnify:
- Contact metagenomics-help@ebi.ac.uk for all enquiries about MGnify.
- Follow @EBImetagenomics to receive the latest news about updates to our services.
- EBI metagenomics tutorials and help pages
Publications:
- White paper: Finn RD, Balech B, Burgin J et al. Establishing the ELIXIR Microbiome Community [version 1; peer review: 1 approve with reservations]. F1000 Research 2024, 13(ELIXIR): 50. (doi: https://doi.org/10.12688/f1000research.144515.1)
- Agafonov A, Mattila K, Tuan CD et al. META-pipe cloud setup and execution [version 1; referees: awaiting peer review]. F1000Research 2017, 6(ELIXIR):2060 (doi: 10.12688/f1000research.13204.1)
- Alexandre A, Mitchell AL, Boland M et al. A new genomic blueprint of the human gut microbiota. Nature 2019 (doi: https://doi.org/10.1038/s41586-019-0965-1)
- Mitchell AL, Scheremetjew M, Denise H et al. EBI Metagenomics in 2017: enriching the analysis of microbial communities, from sequence reads to assemblies. Nucleic Acids Res. 2018; 46: D726-D735 (doi: 10.1093/nar/gkx967).
- Klemetsen T, Raknes IA, Fu J et al. The MAR databases: development and implementation of databases specific for marine metagenomics. Nucleic Acids Res. 2018; 46: D692-D699 (doi: 10.1093/nar/gkx1036).
- Santamaria M, Fosso B, Licciulli F, Balech B, Larini I, Grillo G, De Caro G, Liuni S, Pesole G. ITSoneDB: a comprehensive collection of eukaryotic ribosomal RNA Internal Transcribed Spacer 1 (ITS1) sequences. Nucleic Acids Res. 2018; 46: D127–D132 (doi: 10.1093/nar/gkx855)
- Robertsen EM, Denise H, Mitchell A et al. ELIXIR pilot action: Marine metagenomics – towards a domain specific set of sustainable services [version 1; referees: 1 approved, 2 approved with reservations]. F1000Research 2017, 6(ELIXIR):70 (doi: 10.12688/f1000research.10443.1)
Commissioned Services
The Microbiome Community has been involved in a number of short-term, technical projects called Commissioned Services. These have included: