The Oceans represents the largest biosphere (~97%) on earth. The micro-organisms present within the oceans are responsible for fundamental processes of life, such as nutrient cycling, and produce 50% of the world’s oxygen. However, these organisms are poorly represented in databases.
Marine metagenomics is the study of the genomes of these micro-organisms, as sampled directly from the ocean environment. It is a new and rapidly expanding area of research, and there is a danger that data is produced faster than users are able to analyse, interpret and share it. There is an urgent need to create a data management infrastructure dedicated to marine metagenomics research.
This Community aims to develop a sustainable metagenomics infrastructure to enhance research and industrial innovation within the marine domain.
Services offered
- Marine Metagenomics Portal (MMP): freely accessible microbial genomics and metagenomics reference databases that include MarRef, MarDB, and MarCat. The MMP also includes a search engine that you can use to query all three databases at once.
- MGnify: an automated pipeline for analysing and archiving metagenomic data. You can use a range of services to search and retrieve hundreds of thousands of datasets and billions of annotations.
- ITSoneDB: a database for fungal ITS1 sequences.
Goals of the Community
To develops standards and best practices for the marine domain
- View the first publication on best practices in GigaScience. This is a foundation for a community standard to enable reproducibility and better sharing of metagenomics datasets.
- Driving the use of CWL for the description of metagenomics analysis pipelines to increase transparency and reproducibility of them. You can find examples in the GitHub repository.
To provide databases specific to marine metagenomics
- Marine Metagenomics Portal (MMP): We provide contextual and sequence reference databases via the MMP. These databases include:
- ITSoneDB: a marine fungal ribosomal RNA Internal Transcribed Spacer 1 (ITS1) sequences database.
- Eukaryotic gene catalogue.
To develop tools and pipelines to enhance metagenomics analyses
- META-pipe: We are currently developing the META-pipe pipeline, a free resource for analysing, archiving and browsing metagenomic and metatranscriptomic data, like MGnify.
- MAR BLAST: a search engine for interrogating marine metagenomics datasets. MAR BLAST provides BLAST search on all genes and protein coding sequences from the marine databases MarRef, MarDB and MarCat (see the Marine Metagenomics Portal).
- MGnify, which includes:
- Search service: rich faceted search that allows the discovery of marine datasets.
- Sequence similarity search: allows the querying of millions of peptides, many of which are derived from a marine environment.
- Programmatic interface: allows access to the vast amount of marine metagenomics, metatranscriptomics, amplicon and assemblies (as well as other biomes). For details see the API documentation.
To organise training workshops
- We organise workshops for end users, such as “Metagenomics data analysis” and “Metagenomics bioinformatics”, where training materials and videos are made available. See the Events page to find upcoming workshops.
Example use of MGnify
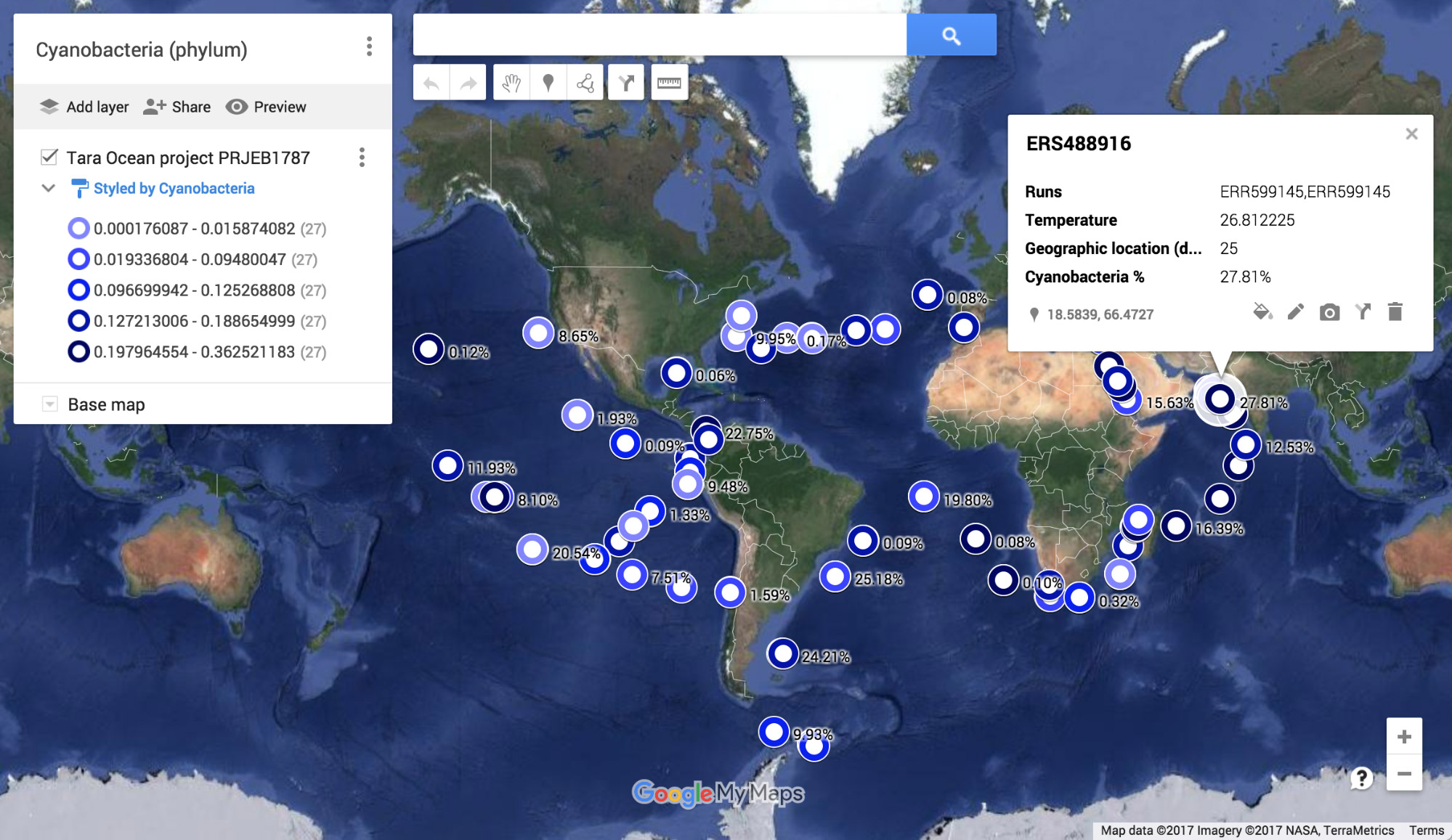

Using the MGnify API, it is possible to access the functional and taxonomic profiles, together with associated sample metadata. This, for example, allows the plotting of the relative abundance of different organisms at different sampling sites (left, map indicating the relative abundance of cyanobacteria at the different sites visited within the Tara Oceans Project). It also allows correlation between species, environmental conditions and biological traits, that are independently evaluated within our pipelines: right, the correlation of photosynthesis (GP:0015979, orange), thylakoid membranes (GO:0009579, blue) and the abundance of the photosynthetic cyanobacteria (green).
Commissioned Services
The Marine Metagenomics Community has been involved in a number of short-term, technical projects called Commissioned Services. These have included:
Leadership

(ELIXIR Germany)

(EMBL-EBI)

(ELIXIR France)

(Communities Coordinator, ELIXIR Hub)
Find out more
- Marine Metagenomics Portal community page
- Contact mmp [at] uit.no for any enquiries about the Marine Metagenomics Portal.
- Contact metagenomics-help [at] ebi.ac.uk for all enquiries about EBI metagenomics.
- Follow @EBImetagenomics to receive the latest news about updates to our services.
- EBI metagenomics tutorials and help pages
- Publications:
- Agafonov A, Mattila K, Tuan CD et al. META-pipe cloud setup and execution [version 1; referees: awaiting peer review]. F1000Research 2017, 6(ELIXIR):2060 (doi: 10.12688/f1000research.13204.1)
- Alexandre A, Mitchell AL, Boland M et al. A new genomic blueprint of the human gut microbiota. Nature 2019 (doi: https://doi.org/10.1038/s41586-019-0965-1)
- Mitchell AL, Scheremetjew M, Denise H et al. EBI Metagenomics in 2017: enriching the analysis of microbial communities, from sequence reads to assemblies. Nucleic Acids Res. 2018; 46: D726-D735 (doi: 10.1093/nar/gkx967).
- Klemetsen T, Raknes IA, Fu J et al. The MAR databases: development and implementation of databases specific for marine metagenomics. Nucleic Acids Res. 2018; 46: D692-D699 (doi: 10.1093/nar/gkx1036).
- Santamaria M, Fosso B, Licciulli F, Balech B, Larini I, Grillo G, De Caro G, Liuni S, Pesole G. ITSoneDB: a comprehensive collection of eukaryotic ribosomal RNA Internal Transcribed Spacer 1 (ITS1) sequences. Nucleic Acids Res. 2018; 46: D127–D132 (doi: 10.1093/nar/gkx855)
- Robertsen EM, Denise H, Mitchell A et al. ELIXIR pilot action: Marine metagenomics – towards a domain specific set of sustainable services [version 1; referees: 1 approved, 2 approved with reservations]. F1000Research 2017, 6(ELIXIR):70 (doi: 10.12688/f1000research.10443.1)
- Slides about the Marine Metagenomics Use Case - from the ELIXIR All Hands Meeting, March 2017.

